A better model for the interpretation of protein hydrogen exchange data
When studying stability and folding of proteins hydrogen exchange substitution is a very important technique. However, the interpretation of the data is complicated by electrostatic effects and the magnitude of these has not been accounted for until now. Mulder et al. have now shown a way to better foretell these effects and thereby provide a key link between experiment and computational modeling of proteins.
Hydrogen exchange (HX) substitution has played a pivotal role in characterizing the globular nature of proteins, the existence of partial unfolding, the hierarchy of folded protein states, as wells as probing the stability of protein structure towards local unfolding.
Unfortunately, electrostatic effects are obfuscating these predictions, especially in studies of intrinsically disordered proteins (IDPs), which limits the utility of HX as a quantitative method to study protein stability, folding, and dynamics.
This situation is about to change, as Dass, Corlianò, and Mulder now publish a way to improve the interpretation of HX data. To highlight this breakthrough, Biophysical Journal featured their study in a ‘New and notable’.
Comprehensive model for calculating intrinsic exchange rates
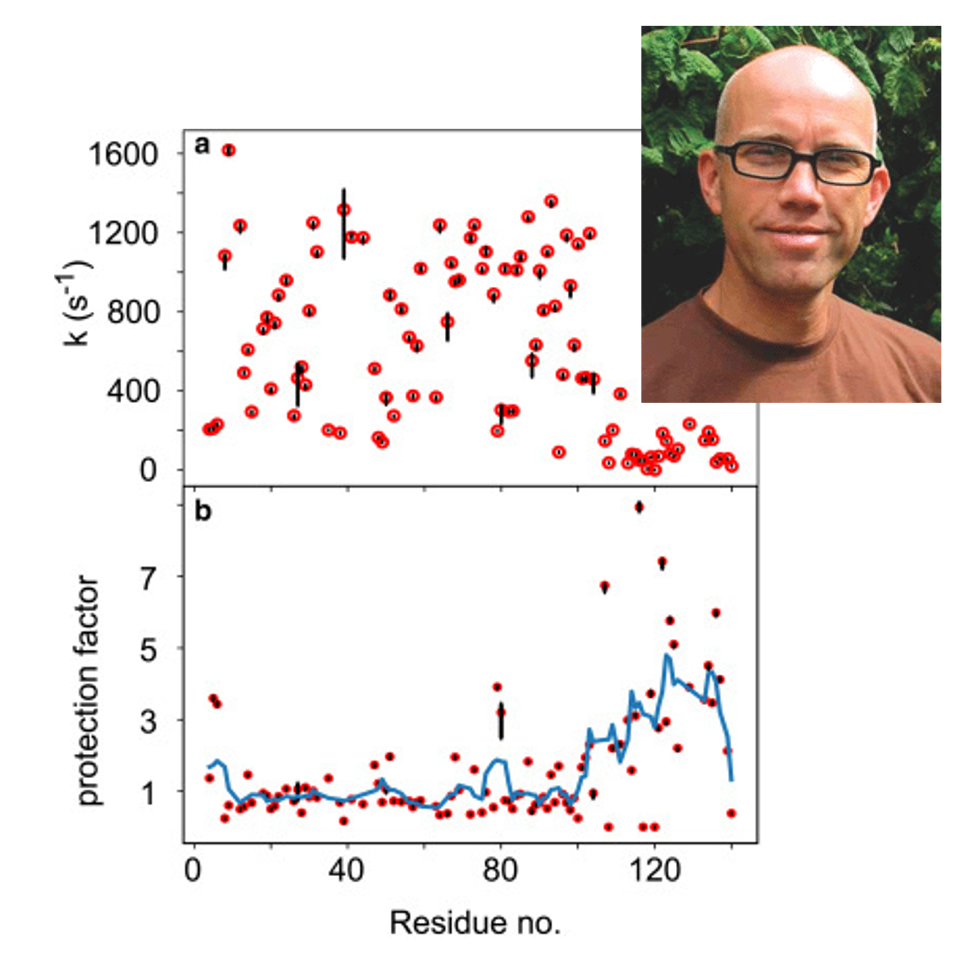
Amide exchange measurements are widely used to characterize protein stability but require information about the unfolded protein state to be accurate. Unfortunately, current approaches only account for the local environment, i.e., take into consideration only the identity of the two side chains that flank the amide bond. Mulder et al. show that it is necessary to account for the more distant environment, as long-range effects are primarily associated with electrostatic interactions. By using the human IDP α-synuclein as a proxy for the unfolded protein state, Mulder et al. show that a hybrid mean-field approach can effectively compute the electrostatic potential at all backbone amide positions along the chain.
They demonstrate that the electrochemical potential provides the missing link between electrostatic theory and HX rates measured in an experiment: A fourfold reduction in hydroxide concentration near the protein backbone is predicted for the C-terminal domain of α-synuclein, a prognosis that is in direct agreement with experimentally derived protection factors from NMR spectroscopy. Thus, impeded HX for this polypeptide region is not the result of intramolecular hydrogen bonding and/or structure formation.
Consequently, the researchers have developed a comprehensive model for calculating intrinsic exchange rates for polypeptide amide backbone hydrogens that can account for electrostatic effects. Being able to account for these effects can aid the correct interpretation of experimental HX protection factors and future HX data will most likely find greater use in the construction of ensemble models for IDPs, with electrostatic calculations providing a key link between experiment and model.
READ ALSO:
- Prediction of protein disorder from amino acid sequence
- Empty spaces, how do they make a protein unstable?
Read more about the results in Biophysical Journal:
The contribution of electrostatics to hydrogen exchange in the unfolded protein state by Rupashree Dass, Enrico Corlianò, and Frans A. A. Mulder.
Toward a proper interpretation of hydrogen exchange data in disordered proteins by Nikolai R.Skrynnikov
The research was carried out by researchers from Interdisciplinary Nanoscience Center (iNANO) and the Department of Chemistry at Aarhus University in collaboration with researchers from University of Florence (IT).
The work was made possible by having access to the NMR spectrometers at the Danish Center for Ultrahigh-Field NMR Spectroscopy (Ministry of Higher Education and Science grant AU- 2010-612-181) at iNANO.
For further information, please contact
Associate Professor Frans A. A. Mulder
Department of Chemistry and Interdisciplinary Nanoscience Center (iNANO)
Aarhus University
Email: fmulder@chem.au.dk