3D Snapshots Unveil the Intricate Dance of RNA Folding
In a groundbreaking development, researchers from the United States and Denmark have successfully captured 3D images of individual RNA nanoparticles in the midst of their folding process. Utilizing a cutting-edge electron microscopy technique, the team has unlocked new insight into the intricate folding dance leading to the final shape of RNA molecules.
The flexibility of RNA makes it notoriously challenging to study, as its structure can shift into numerous forms depending on environmental conditions. Traditional imaging methods, such as cryo-electron microscopy (cryo-EM) single-particle averaging (SPA) analysis, rely on averaging data from thousands of selected molecules with common shapes, making it difficult to capture the unique shapes of individual RNA molecules.
In a study published in Nature Communications, researchers from the Molecular Foundry at Lawrence Berkeley National Laboratory and the Interdisciplinary Nanoscience Center at Aarhus University explored the folding process of flexible RNA molecules. They employed an innovative technique capable of studying the 3D shape of individual molecules without averaging. This technique builds on advanced Individual-Particle cryo-Electron Tomography (IPET), a specialized approach that focuses on single molecule 3D imaging in cryo-preserved samples (see figure and fact box below). Previously, this technique has been used to study how nucleosomes fold DNA and induce phase transitions.
Historically, scientists believed that obtaining 3D images from a single molecule was impossible due to weak signals. "It was considered a dead-end method since 1970s," said Gang Ren, a staff scientist at the Molecular Foundry, who co-led the research alongside Ebbe Andersen from Aarhus University.
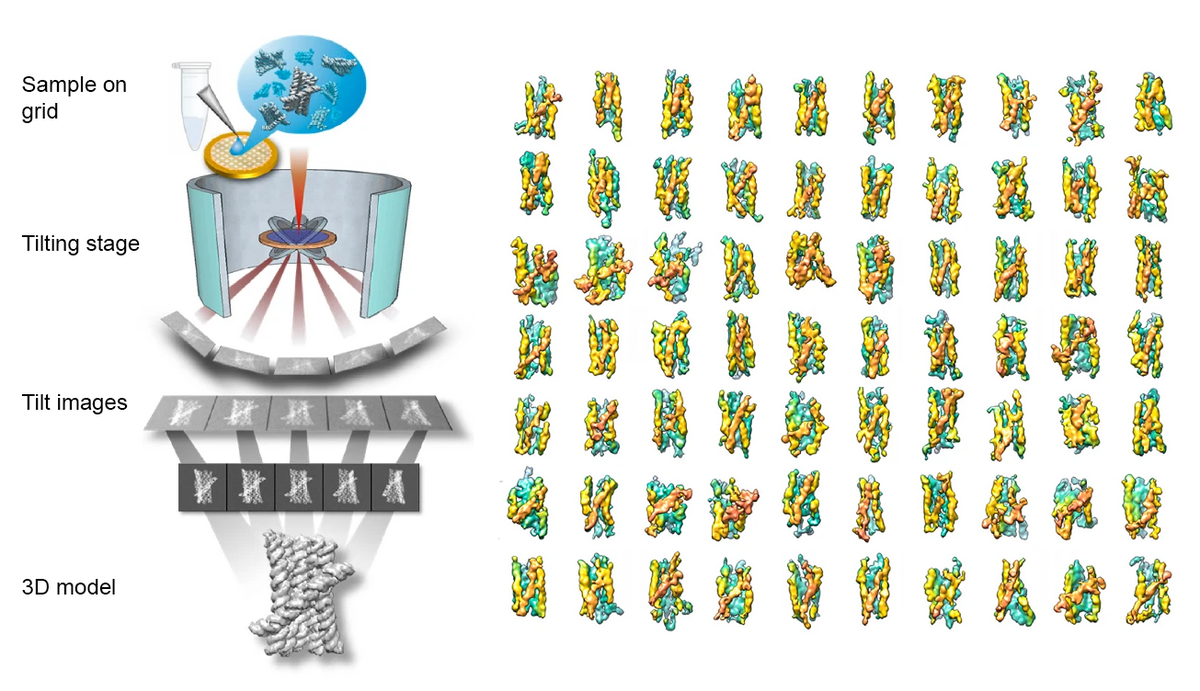
In the current study, the researchers used IPET to study RNA origami – artificially structured RNA molecules engineered to fold into specific nanoscale shapes. Ebbe Andersen and colleagues had previously used the cryo-EM SPA method to study the 3D structure of RNA origami but the folding process remained elusive. IPET allowed the researchers to capture a snapshot of RNA’s folding landscape through capturing molecules in various stages of folding, from immature states to their optimal shape. The researchers were able to observe a folding trap and a shift to a more compact form, enabling creation of a “movie” depicting RNA’s dynamic folding process (see video).
“The IPET technique provides us with a more dynamic view of the molecular world. It is our hope that this insight will enable us to engineer the folding of more effective RNA vaccines and dynamic sensors for molecular medicine”, explains Ebbe Andersen.
Fact box:
Individual-Particle cryo-Electron Tomography (IPET) is a new technique for obtaining the 3D structure of a single macromolecular particle at low to intermediate resolution, without requiring the selection or averaging of images from different particles. This technique incorporates several key advancements, including cryo-electron tomography, focused electron tomography reconstruction algorithms, missing-wedge correction, contrast enhancement, electron dose optimization, and the use of graphene grids. The method enables the visualization of the overall distribution of macromolecular structure in solution. IPET was developed by Gary Ren and Jianfang Liu at the Molecular Foundry, Lawrence Berkeley National Laboratory.
Cryo-electron microscopy (cryo-EM) single-particle averaging (SPA) analysis is a widely used technique for obtaining high-resolution images of biomolecules like RNA, DNA, and proteins. The technique involves freezing the molecules in a glass-like state of ice and imaging them using an electron beam. However, cryo-EM usually requires averaging thousands of homogeneous particles selected from a large pool to build a 3D reconstruction. This requirement makes it difficult to capture dynamic molecules that are continuously changing shape.
About the research
Study type:
Experimental Molecular Biology
External funding:
This project has received funding from the following research grants: the US National Institutes of Health grants of R01HL115153, R01GM104427, R01MH077303, and R01DK042667 (G.R., J.L., and M.Z.); Independent Research Fund Denmark grant 9040-00425B (E.S.A. and E.K.S.M.); Canadian Natural Sciences and Engineering Research Council grant 532417 (EKSM); European Research Council (ERC) Consolidator grant 683305 (E.S.A., C.G., E.K.S.M.), and Novo Nordisk Foundation Ascending Investigator grant 0060694 (E.S.A. and C.G.).
Conflicts of interest:
The authors declare no competing interests.
Link to the scientific article:
"Non-averaged single-molecule tertiary structures reveal RNA self-folding through individual-particle cryo-electron tomography" by Jianfang Liu, Ewan K. S. McRae, Meng Zhang, Cody Geary, Ebbe Sloth Andersen, and Gang (Gary) Ren
https://www.nature.com/articles/s41467-024-52914-1
Contact information:
Staff Scientist Gang (Gary) Ren
The Molecular Foundry
Lawrence Berkeley National Laboratory
Email: gren@lbl.gov
Associate Professor Ebbe Sloth Andersen
Interdisciplinary Nanoscience Center
Department of Molecular Biology and Genetics
Aarhus University
Email: esa@inano.au.dk