Groundbreaking cryo-electron microscopy at Aarhus University reveals the first structures of a protein that maintains cell membranes
Using cutting-edge electron microscopy, researchers from Aarhus University have determined the first structures of a lipid-flippase. The discoveries provide a better understanding of the basics of how cells work and stay healthy, and can eventually increase our knowledge of neurodegenerative diseases such as Alzheimer’s.
All cells are surrounded by a cell membrane that defines and protects the interior of the cell from the outside world. The membrane consists of two layers of fats called lipids, and the distribution of specific types of lipids between the two sides of the membrane is of great importance for the cell to remain healthy and act dynamically. As lipid flippases are a membrane protein, they are located in the cell's membranes, and they are of great importance to the properties of the membrane, where they move lipids from the cell membrane's exterior to the inner layer.
The researchers have studied the lipid flippase Drs2/Cdc50, which is found in yeast cells, where it actively moves lipids from one side of the cell membrane to the other. Similar proteins are found in many variants in humans, where they are also linked to very active processes in brain cells and thus to mechanisms that are central to neurological diseases and dementia, e.g. Alzheimer's disease.
A protein roughly consists of a long chain of amino acids that folds into a three-dimensional structure that is crucial to their function. The experimental determination of the 3D structures of protein molecules requires advanced techniques and is often a great challenge – especially for membrane proteins which are extremely difficult to handle.
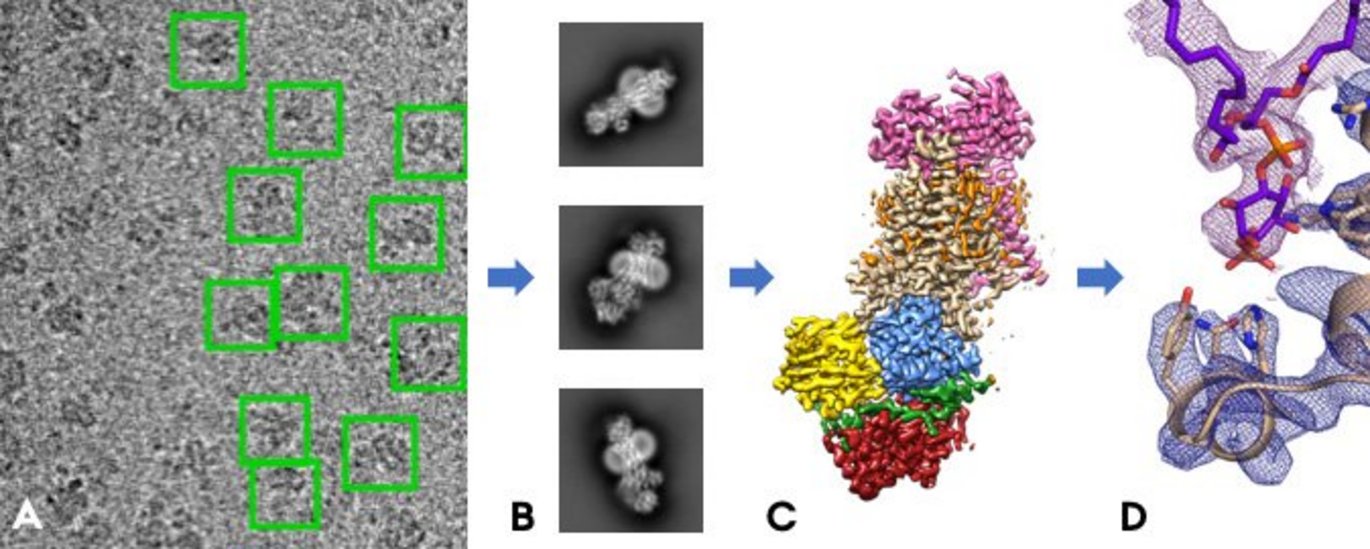
It is not possible to take regular photos of proteins as they are too small, but with the technology of cryo-electron microscopy, a group of researchers from Professor Poul Nissen's research group at Aarhus University have succeeded in determining the three-dimensional structure of Drs2/Cdc50 by taking many hundreds of thousands of "electron photographs" of the molecule. It is the first time anyone has determined a structure from this important family of proteins, the lipid flippases, and the results have just been published in the world-leading journal Nature.
Culmination of over 10 years of work
With the new publication, over 10 years of focused work culminates for a number of students and researchers from Poul Nissen's research group, who also involved collaborative partners at Université Paris-Saclay and the Max Planck Institute of Biophysics in Frankfurt.
"It's fantastic to have been part of the project's final part, see the results fall into place and suddenly be able to explain observations that were detected several years, and which now gives us completely new insights," says PhD student Milena Timcenko, who has been working on the determination of the structure of Drs2/Cdc50 for the past four years together with Assistant Professor Joseph Lyons.
Joseph Lyons adds: ”It’s great to finally to see these membrane protein structures for the first time. The fun begins now to piece together the big picture of how these transport proteins work. It has been an amazing development to be part of.”
Cryo-electron microscopy means that the proteins are cooled down to -170°C in an ultra-thin layer, after which they are "X-rayed" with a powerful beam of electrons and form small shadows on an image chip.
"It's a bit like when you take an X-ray in the hospital, just with electrons instead of X-rays and magnified almost 150,000 times," says Milena Timcenko. "The protein molecules should preferably lie completely motionless to get a sharp image, while the cold temperatures protect the proteins from being destroyed by the powerful electron beam triggered by a voltage of 300,000 volts. That's why we cool them down. The trick is to freeze the samples quickly enough so that no ice crystals are formed, and in a layer that is thin enough that the proteins do not shade one another. We have become quite good at this in Aarhus.”
The results attract huge international international attention
By combining thousands of images where the protein is observed in all possible random directions, one can put together a 3D image that allows to build a model of the protein molecule, atom by atom, thousands of atoms. The model contributes to the basic understanding of how lipid flippases work and can therefore explain some of the diseases that arise from mutations in this family of proteins. This means that small changes have occurred in the 3D structure, and the researchers can now describe the effect of these changes.
This is the first time a completely new membrane protein structure is determined by cryo-electron microscopy in Denmark, and the results attract huge international attention. The work on the large datasets has been made possible by a great effort also to develop research infrastructure in the area, which after several years of work is now in place in Aarhus.
Professor Poul Nissen is very pleased with this development: “With this research infrastructure and strongly growing expertise in cryo-electron microscopy, we are at the forefront of the world competition for new knowledge and opportunities to understand cell biology, drugs and biotechnology at the molecular level. This enables us to maintain and develop Denmark's competitive position in structural biology. We are soon expanding the microscopy facilities with support from the infrastructure program from the Ministry of Research and Innovation and can thus open up access for research groups at all Danish universities, industry and start-up companies. This is essential for our ability to discover new mechanisms in the interaction of physics, chemistry, biology and medicine and to develop new and innovative technologies."
The research has been supported by the Danish Research Council, the Danish National Research Foundation, and the Danish Ministry of Research and Innovation (now called the Ministry of Higher Education and Science), as well as the Carlsberg Foundation, the Lundbeck Foundation and the Novo Nordisk Foundation. Milena’s studies were funded by a European PhD program fellowship from the Boehringer-Ingelheim Foundation.
Link to the scientific article in Nature: Structure and autoregulation of a P4-ATPase lipid flippase: Milena Timcenko1,5, Joseph A. Lyons1,5, Dovile Januliene2,5, Jakob J. Ulstrup1, Thibaud Dieudonne3, Cedric Montigny3, Miriam-Rose Ash1, Jesper Lykkegaard Karlsen1, Thomas Boesen1,4, Werner Kühlbrandt2, Guillaume Lenoir3, Arne Möller2 & Poul Nissen1
1DANDRITE, Nordic EMBL Partnership for Molecular Medicine, Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark. 2Max Planck Institute for Biophysics, Frankfurt, Germany. 3Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Université Paris-Sud, Université Paris-Saclay, Gif-sur-Yvette, France. 4Interdisciplinary Nanoscience Center (iNANO), Aarhus University, Aarhus, Denmark. 5These authors contributed equally: Milena Timcenko, Joseph A. Lyons, Dovile Januliene.
For further information, please contact
Professor Poul Nissen
Department of Molecular Biology and Genetics/DANDRITE
Aarhus University, Denmark
pn@mbg.au.dk – +45 2899 2295
PhD student Milena Timcenko
milena@mbg.au.dk - 3013 4619