New method for building 3D RNA nanostructure ready to knock down target genes
The research groups of assoc. prof. Ebbe S. Andersen and prof. Jørgen Kjems have recently published a paper in Biotechnology Journal describing the design, construction and characterization of an RNA origami with built-in small interfering RNAs (siRNAs).
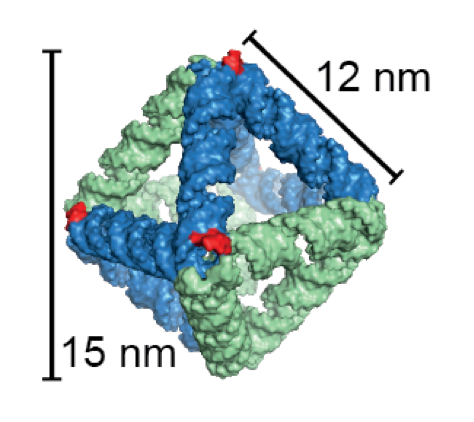
DNA and RNA molecules each have the ability to assemble it-self in two-and three-dimensional structures based on the order of the units the molecules are built of. This feature has been utilized in recent years to program DNA[1] and RNA[2] to form origami structures.
Encouraged by recent applications of RNA nanostructures in nanomedicine Høiberg et al. have developed a unique way to construct a RNA nanodevice with the built-in property to knock down target genes.
The designed RNA nanostructure is an octahedron, which is the first wireframe structure built with the scaffolded RNA origami technique. The octahedron has six intrinsic small interfering RNAs (siRNAs) that can be cleaved out of the structure by the enzyme Dicer. The released siRNAs can knockdown a target gene via the RNA interference pathway. A control structure without Dicer recognition sites were shown to produce significantly less knockdown.
The findings suggest a possible mechanism that could be incorporated into future designs of nanostructures with conditional siRNA release.
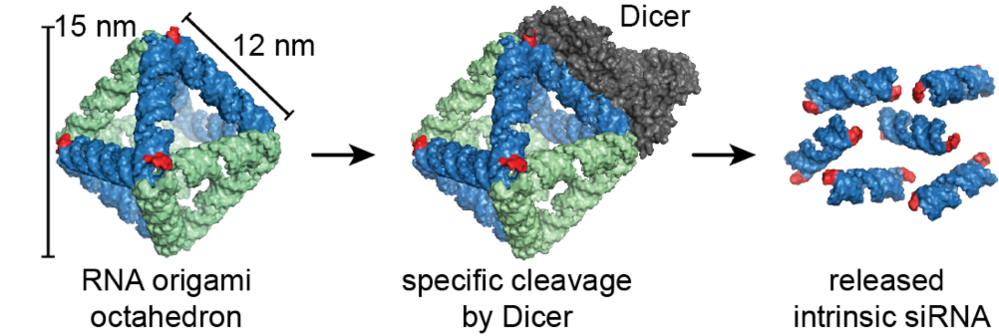
Figure: A 3D scaffolded RNA origami octahedron with intrinsic siRNAs. The enzyme Dicer binds the structure (1) at specific recognition sites and cleaves the structure, which releases the siRNAs (2). The octahedron was demonstrated to efficiently silence a target reporter gene when transfected into H1299 cells.
The project started out as a contribution to the 2011 BIOMOD competition (http://biomod.net), where the Aarhus team "Danish Nano Artists" won the Grand Prize.
Read more about the their successful BIOMOD contribution: http://mbg.au.dk/en/news-and-events/news-item/artikel/inano-students-win-biodesign-competition-at-harvard-university-1/).
Read more about the research performed in the Andersen Lab for Biomolecular Design at andersen-lab.dk.
The research has been carried out by researchers from Aarhus University (Department of Molecular Biology and Genetics and Interdisciplinary Nanoscience Centre). Assoc. prof. Ebbe S. Andersen and professor Jørgen Kjems, Aarhus University, have been in charge of the research team behind the study just published in the journal Biotechnology Journal.
An RNA origami octahedron with intrinsic siRNAs for potent gene knockdown. Hans Christian Høiberg, Steffen M. Sparvath, Veronica L. Andersen, Jørgen Kjems*, Ebbe S. Andersen*. doi: 10.1002/biot.201700634
This work was supported by the Danish National Research Foundation and the Danish Council for Independent Research.
For further information, please contact:
(Photo: Lars Kruse, AU Photo) |
(Photo: Jesper Rais, Rais Foto) |
Assoc. prof. Ebbe Sloth Andersen Interdisciplinary Nanoscience Center Tel: +45 41178619 | Professor Jørgen Kjems Interdisciplinary Nanoscience Center Tel: +45 28992086 |
References:
[1] P. W. K. Rothemund. 2006, 440, 297
[2] P. Guo, Nat. Nanotechnol. 2010, 5, 833

